Renkawitz Lab - Cell & Mechano Biology of Immune Responses
Research Concept & Vision
Immune Cells to Discover Novel Principles in Cell- & Mechano-Biology. The immune system consists of an elaborate orchestration of cell types specialized for different molecular processes. Due to the spectrum of these specialisations, immune cells represent an illuminating cellular model to identify general principles in cell- and mechano-biology. We employ immune cells to unravel fundamental principles and mechanisms in cell motility & navigation, tissue microenvironments, organelle positioning & organelle mechanobiology, tissue surveillance by macropinocytosis, and host-pathogen interactions.
Our lab interdisciplinary combines advanced live-cell microscopy, image analysis, genetic engineering by CRISPR, custom-made engineering of micro-environments by microfluidics and 3D collagen matrices, and unbiased system-wide approaches like genome-wide CRISPR screening, transcriptomics, and proteomics. Thereby, we aim to discover and unravel fundamental principles in the cell- and mechano-biology of the immune system and their misregulation in disease.
General questions
(i) How does the extracellular microenvironment regulate cellular behaviour?
(ii) How do organelles achieve their intracellular position and mechanical stability in cells?
(iii) How do pathogens hijack the principles of the cell-to-matrix interplay?

Research Topics in the Lab
Cell Motility & Navigation
Immune cells are fast-moving cells that must navigate through crowded three-dimensional environments (Kameritsch & Renkawitz, Trends in Cell Biology 2020). While some cell types, like mesenchymal cells, proteolytically break down obstacles as they move, many immune cells, like dendritic cells and T cells, typically migrate without remodelling or digesting their local surrounding. If these immune cells were to generate tunnels through the body by local digestion, they would end up perforating and thus damaging the local tissue microenvironment.
We aim to unravel the mechanobiology and cell biology of how immune cells efficiently migrate and navigate their path. Thereby, we discovered that immune cells use their nucleus as a ruler to probe their surroundings for the largest pores and thereby migrate along the path of least resistance (Renkawitz et al, Nature 2019). Further, our research has identified that immune cells efficiently adapt their migratory path between dominant mechanical and chemotactic guidance cues, which involves a switch in their cell polarity and effective repositioning of the nucleus through a process named nucleokinesis, driven by myosin-II mediated contractility (Kroll et al, EMBO J 2023). Moreover, we discovered that the intracellular parasite Toxoplasma gondii hijacks the myosin -mediated contractility of migrating immune cells, facilitating its movement and transport through complex and confining microenvironments (Ruiz et al, bioRxiv 2024).
Selected Publications:
Ruiz et al, bioRxiv 2024
Kroll et al, EMBO J 2023
Holtkamp et al, Nature Immunology 2021
Kameritsch & Renkawitz, Trends in Cell Biology 2020
Renkawitz et al, Nature 2019

—————————————————————————————————————————————————————————————————————————————————————————————————
Organelle Positioning & Integrity
Cells and their organelles are continuously exposed to forces, either from the microenvironment like fluid flows in the blood vascularity, contraction in the beating heart, stiffness gradients and confining extracellular matrices in tissues, or by their dynamics generated by cytoskeleton forces.
We employ immune cells as a cellular model that encounters extensive mechanical stresses as they traverse tissue, to understand how organelles maintain their intracellular positioning (Kroll & Renkawitz, EMBO reports 2024) and their mechanical stability. Our research has discovered that the centrosome, a non-membrane surrounded organelle, is susceptible to mechanical breakage when cells navigate through tissue environments, causing the emergence of two competing microtuble-organizing centers (MTOCs) and cellular entanglement in the local surrounding (Schmitt et al, Science Advances 2025).
Selected Publications:
Schmitt et al, Science Advances 2025
Kroll & Renkawitz, EMBO reports 2024

—————————————————————————————————————————————————————————————————————————————————————————————————
Cytoskeletal Dynamics & Adaptability
Almost all moving cells employ the actin cytoskeleton as an intracellular force generator to move the entire cell body forward. In addition, cells locally extend protrusions in an actin-dependent manner to engulf solid particles by phagocytosis or extracellular fluid by macropinocytosis.
Using macrophages and immature dendritic cells, our research has discovered that extracellular fluid sampling by macropinocytosis is regulated by the adhesiveness of the local microenvironment (Braun et al, ResearchSquare Preprint 2024). Moreover, in earlier research, we discovered that amoeboid cells (such as dendritic cells) adapt their actin cytoskeleton dynamics to the adhesiveness of the migratory substrate (Renkawitz et al, Nature Cell Biology 2009). Thereby, we could show that not tracks of adhesive substrates but gradients of chemoattractants dictate the path of amoeboid cells, endowing these cells with extraordinary flexibility and enabling them to traverse almost every type of tissue (Renkawitz & Sixt, EMBOreports 2010).
Selected Publications:
Braun et al, ResearchSquare (preprint) 2024
Hons et al, Nature Immunology 2018
Renkawitz & Sixt, EMBO reports 2010
Renkawitz et al, Nature Cell Biology 2009
—————————————————————————————————————————————————————————————————————————————————————————————————
Tissue Microenvironments & their Engineering
Whereas in vivo experiments are truly physiological, they do not allow for precise manipulation of environmental parameters and are elaborate for the discovery of detailed molecular mechanisms. In contrast, in vitro experiments on two-dimensional (2D) substrates (e.g. cell culture dishes) enable faster manipulations, but increasing knowledge points to substantial differences in cellular mechanisms in 2D and three-dimensional (3D) environments.
To bridge this gap, we and others developed micro-engineered tissue-mimetic assays to combine the advantage of precise manipulations in 2D assays with the presence of complex 3D microenvironments. Specifically, we implemented the methodology of micro-engineered ”pillar forests’ to study cell migration in vitro in 3D with precisely defined microenvironmental parameters (Renkawitz et al., Methods in Cell Biology 2018). Shortly, these devices provide a 3D migration environment made of PDMS (polydimethylsiloxane), in which two closely adjacent surfaces are interconnected by micron-sized structures such as pillars (or microchannels). Thereby, these devices represent a flattened approximation of complex 3D environments with the advantage of cellular confinement in one plane (XY) close to the imaging surface, enabling high-resolution single-cell imaging.
To discover how extremely fast immune cells navigate through the dense meshwork of interstitial tissue fibres without harming other cells, we developed these environments to represent obstacle courses for immune cells in reconstituted environments (Kroll et al, Current Protocols 2022). Moreover, we employed and further developed the under-agarose assay to mimic cellular confinement by their local microenvironment (Clausen et al, Eur J Immunology 2022). Overall, we complement these assays with experimentation in collagen matrices, ex vivo tissue explants, and in vivo approaches.
Selected Publications:
Kroll et al, Current Protocols 2022
Clausen et al, Eur J Immunology 2022
Renkawitz et al, Methods in Cell Biology 2018

—————————————————————————————————————————————————————————————————————————————————————————————————
Genetic Engineering of Immune Cells
Current knowledge on the molecular mechanisms and cell biological principles of immune cell biology (such as cell migration) is often based on studies utilizing knockout mice, which however hampers screening of large numbers of candidate components due to its time and resource consuming nature. To circumvent this bottleneck, conditionally immortalised hematopoietic precursor cells (Hoxb8 cells) with myeloid and lymphoid potential have been established by the Häcker Lab (Redecke et al, Nature Methods 2013). We recently contributed in showing that Hoxb8 cells can be differentiated into migratory DCs functionally indistinguishable from their primary counterparts (bone-marrow derived DCs) (Leithner et al, Eur J Immunol). Importantly, Hoxb8-FL cells can be efficiently targeted via CRISPR mediated gene editing (Leithner et al, Eur J Immunol). Thus, we employ immortalised hematopoietic precursor cells (Hoxb8 cells) as an accessible cellular model for genetic engineering, including genetic knockouts and fluorescent tagging.
Selected Publications:
Leithner et al, Eur J Immunol 2018
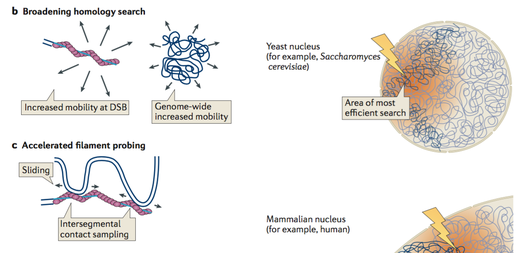
Earlier Research on DNA Repair
During his PhD research in the Lab of Prof. Stefan Jentsch, Jörg visualised homology search during DNA double-strand break (DSB) repair in vivo, using genome-wide analysis of chromatin immunoprecipitation of DSB repair factors in yeast. This showed that homology search is strongly influenced by the chromosomal architecture and nuclear organization (Renkawitz et al, Mol Cell). These findings led to a model, in which homology search during DSB repair proceeds by an accelerated random probing mechanism guided by genomic proximity (Renkawitz et al, Nat Rev Mol Cell Biol).
Selected Publications:
Lademann et al, Cell Reports 2017
Renkawitz, Lademann & Jentsch, Nature Reviews Molecular Cell Biology 2014
Renkawitz et al, Molecular Cell 2013